There is a wealth of information in scientific articles. Europe PMC contains millions of life sciences articles. Finding specific evidence within such a huge corpus of research literature has historically been very challenging for researchers and biocurators.
When I joined the Europe PMC team they had already put in place text mining algorithms to automate the extraction of biological terms and concepts from unstructured text in article abstracts and full text. For example genes, proteins, diseases, organisms, chemicals and biological processes and functions, such as gene-disease relationships or protein-protein interactions.
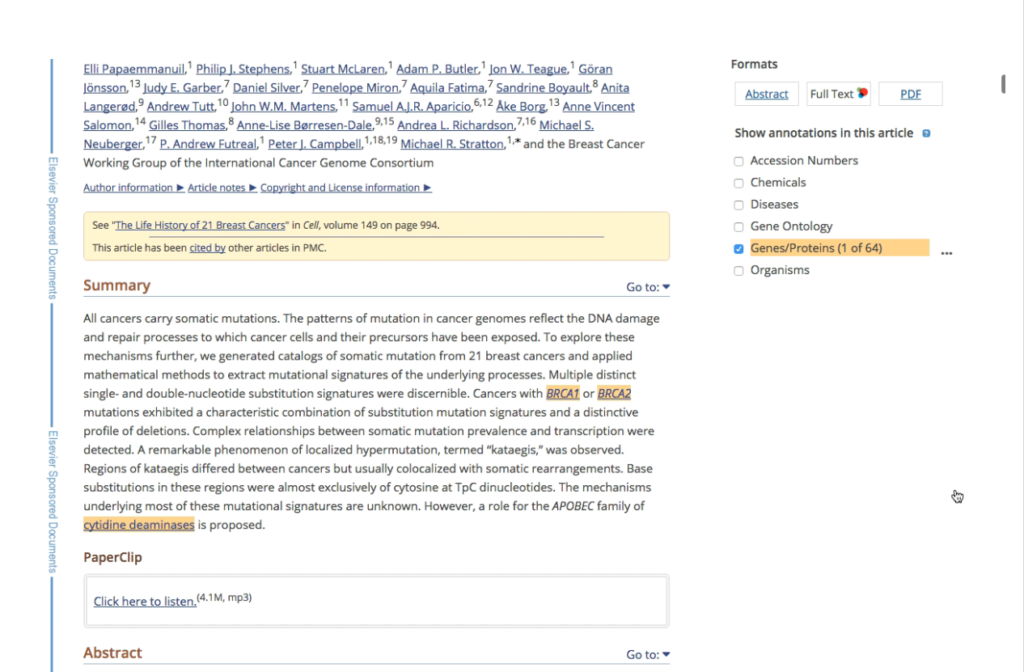
The terms and concepts extracted from the articles were presented in a list in a tab on the article page, rather than in context where they appeared in the article. Annotations were only highlighted on the text of the article abstract (a short summary at the beginning of the article).
The challenge
Biocurators extract evidence from articles to curate and enter into structured databases, such as Uniprot (a protein database) or Flybase (a model organism database). Finding that information can be challenging.
“I was looking for the cellular location (cytoplasm or nucleus) of ribonucleotide reductase. It’s like a needle in a haystack.”
Researchers also have an interest in being able to find relevant articles to their field of resarch that mention particular genes, proteins or mentions of diseases that are the focus of their research.
Our goal for the project was:
To help researchers find useful information in articles quickly, and link to related data resources
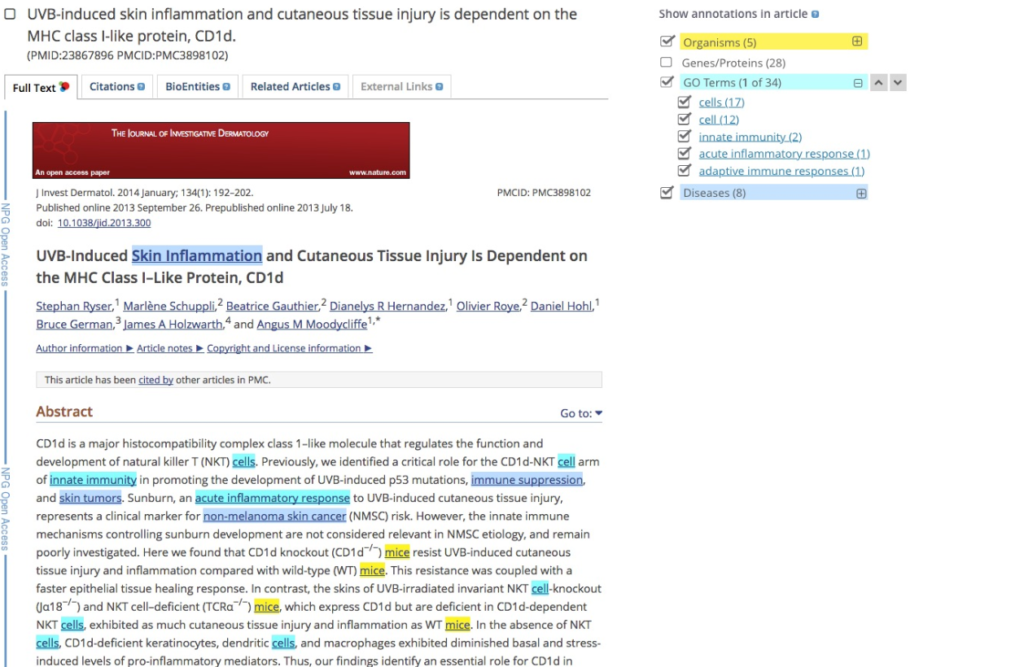
We set out to design an interface that would overlay and highlight annotations on the article text, making it easier for biocurators and researchers to see annotations in context and to understand at a glance if the article was relevant to them. We also wanted to link the annotations to structured data, for example a protein record in Uniprot.
My role as a UX Researcher and Designer
My first task was to understand the user needs more deeply. I started with generative research. My research goal was:
- To understand how researchers and curators find literature and make decisions about what to read
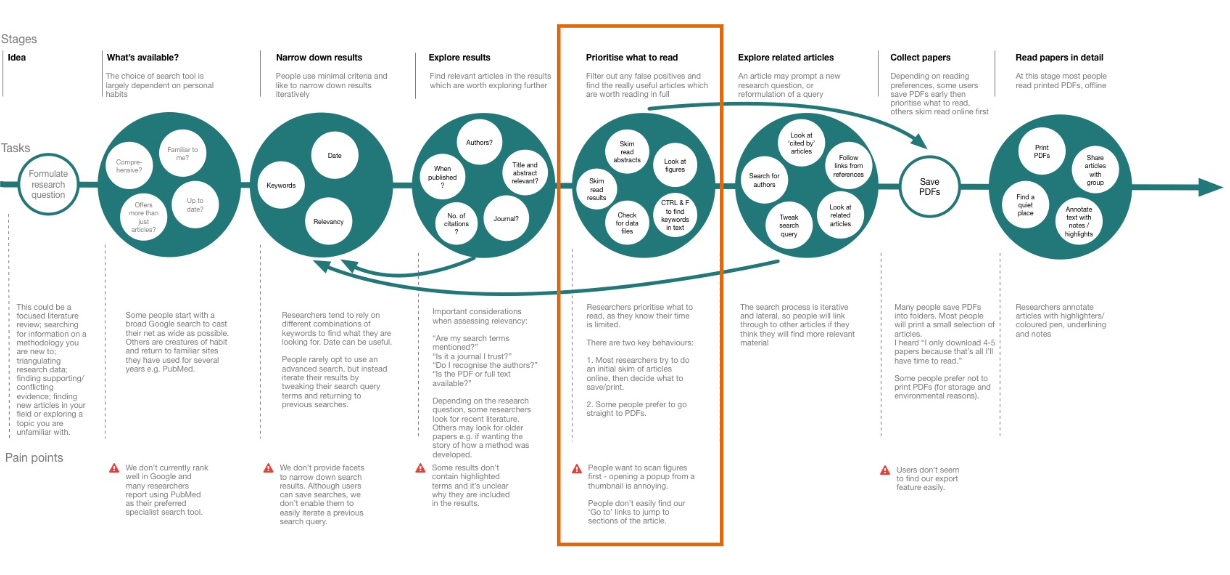
I conducted interviews with 8 researchers and 2 curators. I put together a task model to illustrate how users find evidence to answer a research question.

The steps in the model included:
- Formulate a research question
- Choose a search tool and strategy
- Narrow down results
- Explore results
- Prioritise what to read
- Explore related articles
- Collect articles
- Read articles in detail
Researchers prioritise what they want to read, as their time is limited. The annotations feature aimed to solve user problems in this step of the model. Researchers and curators use different strategies to identify articles which are worth reading in full. These strategies included skim reading the results section and using CTRL & F to find keywords in the text.

Following the user research I developed a prototype to test with users. After one round of testing, we realised that context is everything when it comes to annotations. It was difficult to get good insights without using real annotations data. The developers then created a prototype with real data behind it and we tested it with users. In total we tested with 17 users.
Some of my research questions included:
Do participants discover/use the feature?
- How easy is it to use/navigate through annotations?
- Do users trust the information?
- How do they feel about inaccurate annotations?
- Would they provide feedback if they had the opportunity?
The usability sessions were attended by developers and text miners, so that they could observe first-hand the issues that users faced.
The main findings related to:
Granularity of the annotations
Some terms appeared too frequently, or were too general to be useful e.g. “cell” or “formation”.
“If it’s not specific enough, I end up with a lot of things being highlighted.”
Trust
Users lost trust in the information if there were false positives e.g.
- “oxide” is not an organism, but “oxidae” is
- “ubiquitin” is a process not a gene/protein
“I guess false positives automatically make me anxious about whether to believe…”
Discoverability and engagement
We faced some challenges with making the annotations functionality discoverable and encouraging users to engage with it.
We had some constraints, for example we could only show annotations on articles with open licenses. And we were unable to indicate how many annotations were available due to a performance impact on page loading.
Navigation
We had a panel on the right-hand side of the article where users could turn annotation categories on and off. We discovered that users expected to see a list of terms under the category heading.
“If you click on organisms I’d expect it to expand out and see the unique items e.g. zebrafish”
Users also expected to jump straight to terms in the text and navigate through them.
Outcomes
We launched the annotations feature in 2016 and had plenty of positive feedback about it. But we were aware that there were some usability issues that would be hard to resolve due to the technical and license constraints.
Lessons learned
We learned some really valuable lessons from the project.
- User research and feedback are essential for improving text mining pipelines.
- A prototype with real data provided better user research insights as the accuracy of information and context was key to how well the feature worked for users.
- Trust is quickly eroded when an annotation that has been highlighted doesn’t make sense in context. So providing a way for users to provide feedback to help the team improve the text mining algorithm was essential.
- Compromises between technical / performance constraints and user needs are inevitable – but it’s important for developers, text-miners and UX designers to make design decisions together.
Footnote
Since releasing the annotations feature it has been through two redesigns and had some functional improvements:
- An annotations API was developed in 2017 to provide programmatic access to annotations. I was involved in defining the API methods based on my existing understanding of user needs.
- A redesign in 2019 dealt with the issue of scalability and handling an increasing number of annotation types.
- The accuracy of the annotations was greatly improved by replacing the text mining pipelines with machine learning models.
- Some new functionality was introduced in 2023 to display annotations from supplemental files as well as the full text of articles.
I was involved in these later iterations of the design and functionality as a Product Manager rather than a UX Designer.
References
Designing with algorithms – how text miners and UX can work together, Michele Ide-Smith and Jee-Hyub Kim, UX Cambridge 2016
Annotations page on Europe PMC
Scilite Annotations, Europe PMC team, 9 Sep 2016
Venkatesan A, Kim J, Talo F, et al. SciLite: a platform for displaying text-mined annotations as a means to link research articles with biological data. Wellcome Open Res; 2017. DOI: 10.12688/wellcomeopenres.10210.2.